Below is my previous mentoring experience.
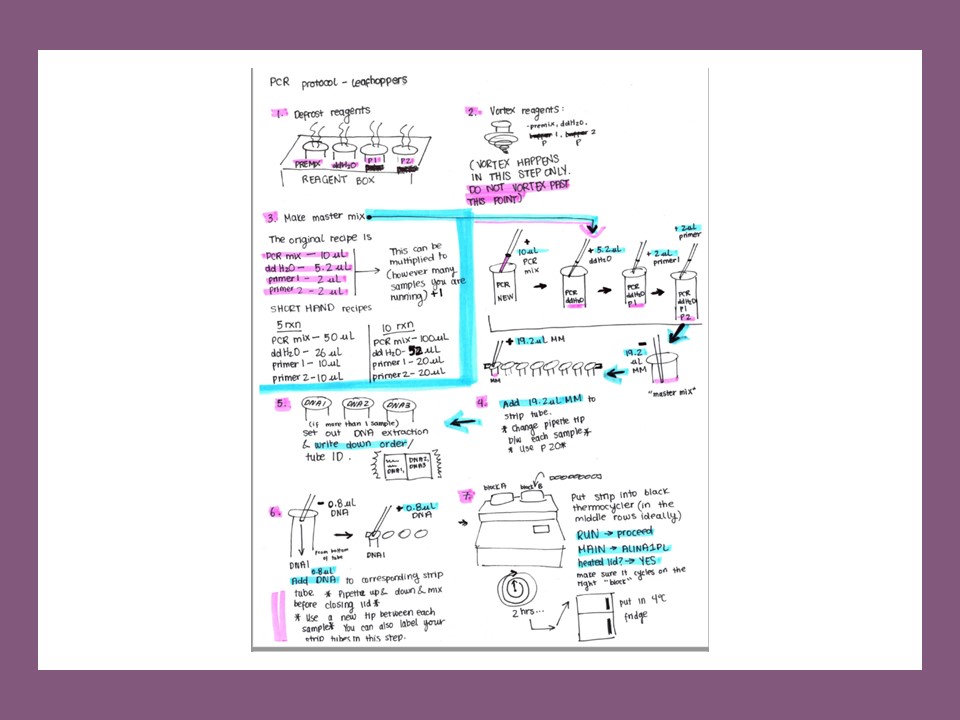
I’ve been mentoring graduate, undergraduate, and high school students in various research projects which involved insect diet analysis using molecular biology techniques. During my postdoctoral work in Dr. Bill Lamp’s lab, I proposed, developed and managed DNA barcoding flow for our lab research: I set up DNA extraction, PCR, and electrophoresis protocols, proposed and acquired reagents and equipment, set up sequencing software, managed sequencing service, trained and coordinated lab members. This DNA barcoding work became the main part of several ongoing projects in Dr. Bill Lamp’s lab, such as: wetland-stream connectivity; dragonflies diet composition; and host plant usage of potato leafhoppers and the spotted lanternfly. Here are the research protocols which we were using.
I’ve been training the students in tissue preparation, DNA isolation, PCR, visualization results using gel electrophoresis, sequence analysis and depositing the sequences to the NCBI GenBank datase, as well as retrieving data from published studies as part of our planned systematic review on molecular approaches to study insect diet. The students work to identify insect host plants, confirm their ingestion, and explore insect host plant switch during insect development. The students also conduct sequence analysis, and learn how annotate the sequence and deposit it to GenBank.
All this work has resulted so far in multiple student GenBank submissions, 2 student school symposium presentations (with 1 award), and two published manuscripts with two of my mentees as coauthors. I enjoy working with my students and seeing their progress, and I’m very happy for their big and small successes.
Over the past 4 years, I mentored several students in different aspects of the molecular gut content analysis of the spotted lanternfly. Jessica and Nurani, our former lab managers, conducted DNA extractions and PCR amplification of a portion of trnL-intron from ingested plants; Darsy, a Ph.D. student, helped with obtaining and depositing plant sequences to the NCBI GenBank database; Hannah, an undergraduate student, was working on meta-barcoding of the lanternfly gut contents using a NGS-approach; Cameron, a research assistant, is currently focusing meta-barcoding of the spotted lanternfly gut contents, sequence analysis, and phylogenetics of the lanternfly host plants.
Hannah, my former mentee and a great helper, made an incredible progress: after less than a month of training she independently conducted all the steps from DNA isolation, PCR, and preparing samples for NGS sequncing. Hannah was working on identification of obtained sequences, as well as a qualitative analysis of the spectrum of ingested host plants. She was also actively participating in another project, a systematic review of molecular approaches to decipher insect herbivore diet (manuscript is published with Hannah as a coauthor).



03/21/21: Our systematic review on choosing an effective PCR-based approach for diet analysis of insect herbivores is published in Journal of Economic Entomology! Hannah is a co-author - so exciting!
04/09/20: To date, my mentees (in bold) have succesfully submitted to GenBank a total of 10 sequences for the chloroplast trnL-gene. All of them have been approved and are already available in the NCBI GenBank database:
Four of these sequences were obtained by my former mentee, Bryan, a high school student!
Bryan, a high school student, was working with us during last Spring semester, and I was really impressed by his incredible progress during those several months. Bryan joined us in January knowing nothing about wet lab techinques for DNA barcoding process; he worked once a week only learning all the steps; and by the end of April he had successfully extracted genomic DNA from 100+ leafhopper samples, amplified plant DNA from leafhopper guts and succesfully obtained and deposited four plant sequences. His work was the essential part of our paper on the use of molecular markers for plant DNA detection from leafhopper gut contents. My other mentee on this project, Nurani, our lab manager at that time, has succesfully set up and conducted feeding experiments with the potato leafhopper (see below); she is a co-author on this paper. My new mentee, Anya, a high school student, has recently started applying a meta-barcoding approach to the bulk samples of the potato leafhopper, to identify all the ingested plants.

12/01/21: My new mentee, Anya, has submitted an abstract for her poster presentation “Using molecular gut content analysis to characterize PLH (Empoasca fabae) movement within a farmscape” for the upcoming meeting of the Entomological Society of America, Eastern Branch (in Philadelphia, PA)
11/04/20: Our paper on detecting ingested host plant DNA in potato leafhopper was published online today in Journal of Economic Entomology! So exciting!
09/23/19: I was very pleased to learn recently that Bryan was admitted to the University of Maryland where he continues studying Molecular Biology. Bryan was also featured in The John Carroll School News. I’m so happy for him!
This was a long-term project in Dr. Lamp’s lab. Using isopods from multiple wetland sites and streams in Maryland Delmarva Bays Wetlands we explored the potential connectivity between wetland and stream communities. Morphological identification of isopod species is tricky, and DNA barcoding is very helpful for estimating how ‘close’ wetland isopod species are to the isopod species inhabiting streams. I mentored three students in this DNA barcoding work, and together with my mentees we isolated and sequenced a portion of mitochondrial gene, cytochrome oxidase 1 (CO1), created a reference library of these sequences, and using a BLAST search engine in the NCBI GenBank database we assessed species and genera identity of isopods. We were able to identify seven unique stream species and four unique wetland species of isopods. We didn’t find an overlap between species which suggests presumably isolation of stream and wetland populations of isopods and may potentially provide an evidence that stream and wetland arthropod communities do not overlap as well. We are currently working on reconstruction of phylogenetic relationships between these isopod species.
One of my mentees, Nina, a high school student, was working on this project more than year. During her work she created a reference library of mitochondrial sequences from stream isopod species - she then ‘BLASTed’ sequences from wetland isopod individuals against this library to determine the matches and estimate how ‘close’ wetland species were to stream species.


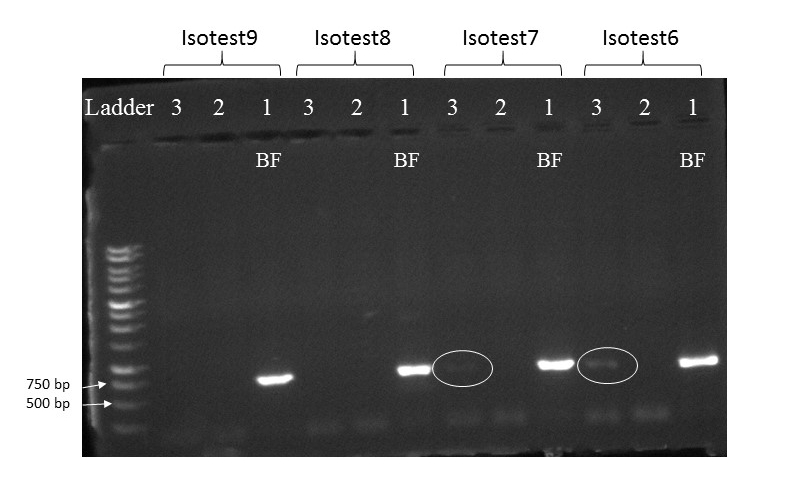
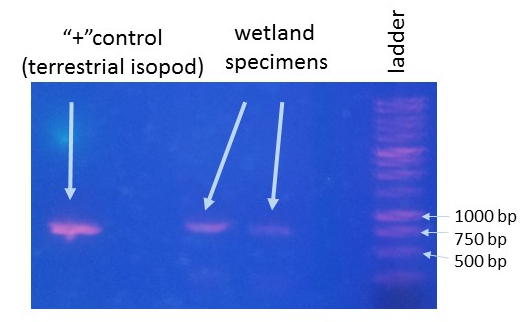
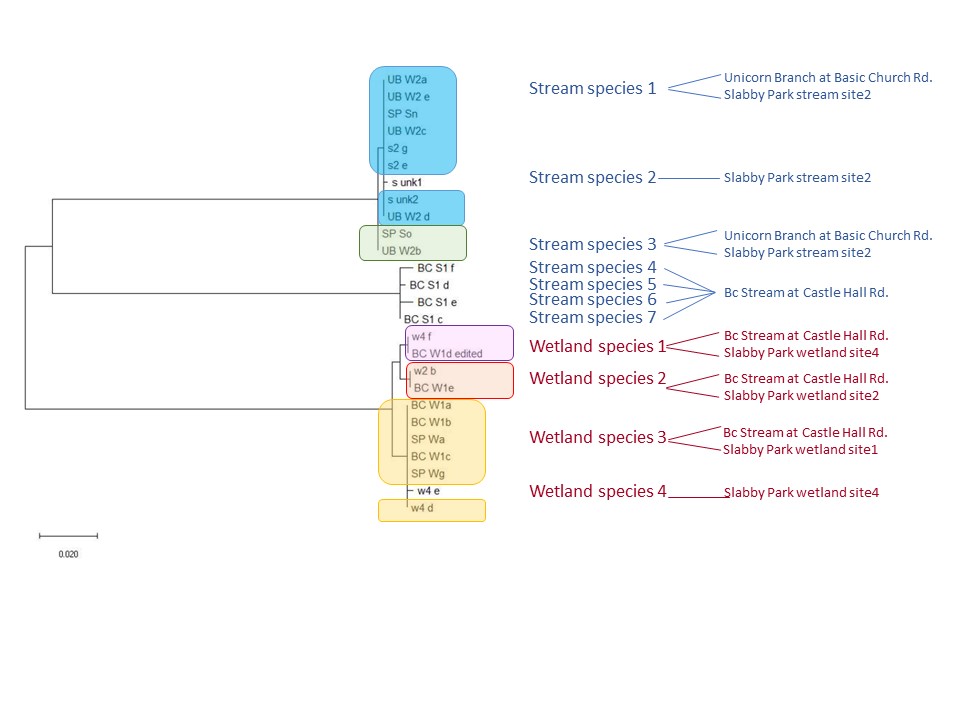
10/06/19: We are currently exploring phylogenetic relationships between stream and wetland isopod species.
05/25/19: Nina was admitted to the University of Maryland where she will study Environmental Science and Policy. Very exciting! She is continuing, however, her work on DNA barcoding of isopods this summer, learning and improving the phylogenetic reconstructions for our isopod samples.
04/26/19: Nina succesfully presented the final results of her year-long project at her school’s research symposium!
02/22/19: Nina, my mentee, a high school student, who was working on this project since September 2018, presented her research project earlier this week at her school’s science fair and got 3rd prize! So exciting!
This was a new challenge in our DNA barcoding work - we needed to amplify DNA from dragonfly prey items. For this, together with my mentees, Maggie and Leela, we successfully amplified and sequenced COI partial gene from dragonfly feces produced during the first 2 hours post ingestion. The sequence analysis with subsequent BLAST results showed that the prey item was a crane fly. Surprisingly enough the DNA from the crane fly wasn’t degraded and an isolated fragment of 667 bp showed good sequence quality. These exciting results can make identification of dragonfly prey possible and accurately confirm (or even point to new) trophic interactions in dragonfly natural habitats.

10/1/20: We’ve started conducting meta-barcoding of Odonata feces using a NGS approach.
06/10/19: DNA from mayfly was detected in feces produced during first 7 hours post ingestion by the same dragonfly individual. The sequence had lower quality but it was still readable.
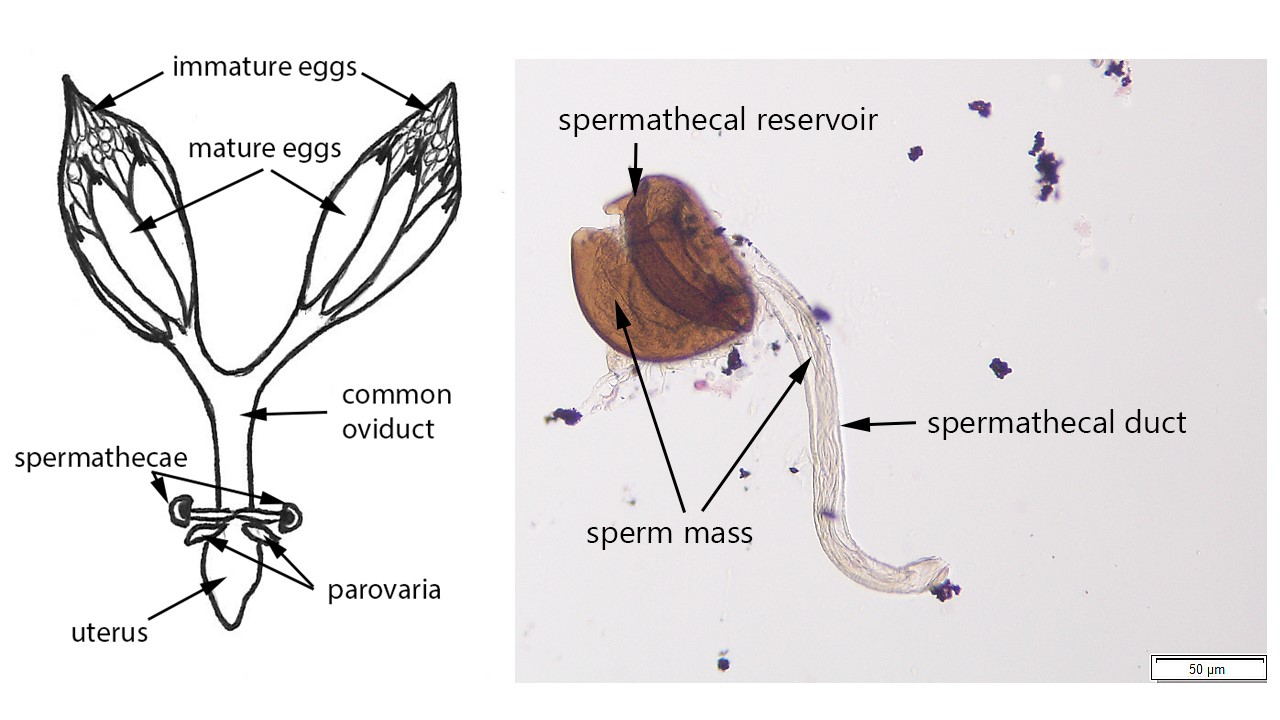
During my postdoctoral work at the University of Wisconsin-Madison I trained a high school student in tissue dissection and isolation, slide preparation, tissue and cell identification, morphological analysis. Claire did a research project on the spotted wing drosophila through the Biotechnology Youth Apprenticeship program. Claire learned how to use a new protocol I developed for dissecting fly tissues and determining fly mating status. She successfully applied this new technique to different fly specimens; she also created beautiful illustrations for an invited manuscript which we published later in Insects. Here are some of the figures Claire made for our paper:
Additionally, Claire compiled a large dataset for temperature and humidity data we used in another paper on the effect of temperature and humidity on the seasonal phenology of the spotted wing drosophila.
As a PlantingScience fellow, during summer/fall 2017 I participated in the PlantingScience program (Botanical Society of America; Digging Deeper project). I was an online scientist mentor and liaison for student research teams (high school and middle school students).
That was my first year with the PlantingScience program. I really enjoyed our summer workshop in Colorado Springs and meeting with biology teachers from around the country, as well as our wonderful trip to the Garden of the Gods. Last Fall, I was a scientist mentor for nine research teams and a liaison for a biology teacher and three research teams in her class. The student teams which I mentored, focused on The Power of Sunlight module and conducted their independent research projects on plant photosynthesis and respiration.
Since then, I stayed with PlantingSceince, and I’ve been serving as a scientist mentor every semester. During Fall 2018, I worked as a member of Master Plant Science Team serving as a liaison for one class and a mentor for three student teams. Great experience! I’m continously looking forward to being a part of this interesting program every semester!



In 2003, at Herzen State University, I started teaching a course on Animal Ecology. Since then, I’ve supervised the research of several undergraduate students, including short projects (as a part of their independent research), as well as their final theses (a Diploma in Russian academia) …more information






Photo credit: Alexandr Mogilev. Thesis project: Ecological adaptations of the Felidae family: example of the Siberian tiger, Pantera tigris altaica (Leningrad Zoo, St. Petersburg, Russia, 2005)